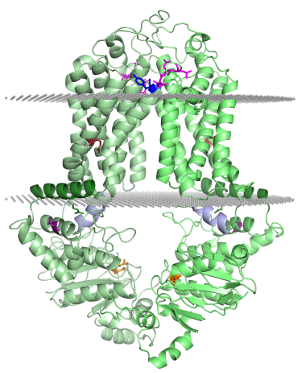
Equilibrium simulations show that uric acid, a substrate of ABCG2 visits Site 2, which was identified by Laszlo et al and involves R482:
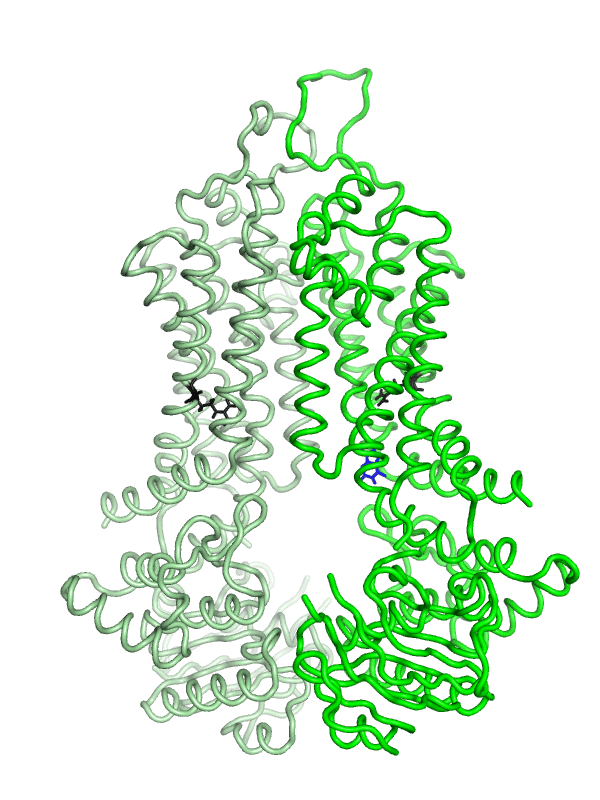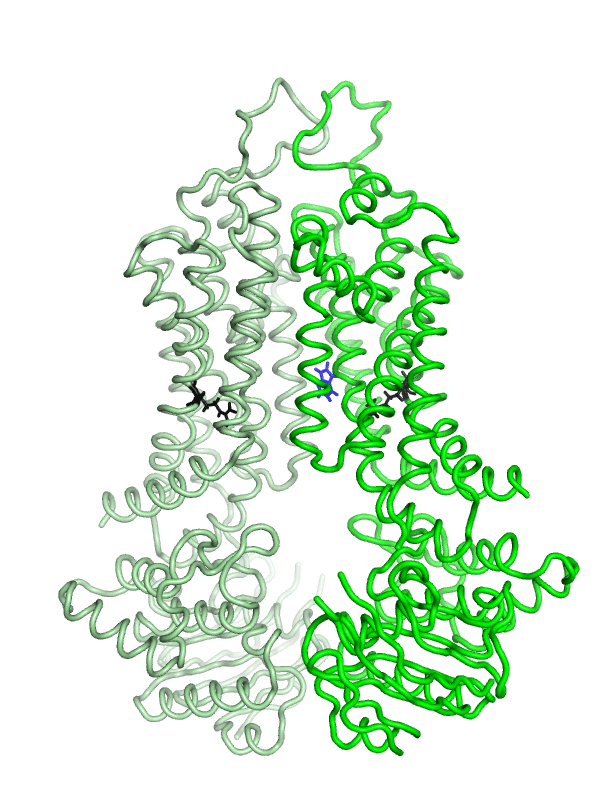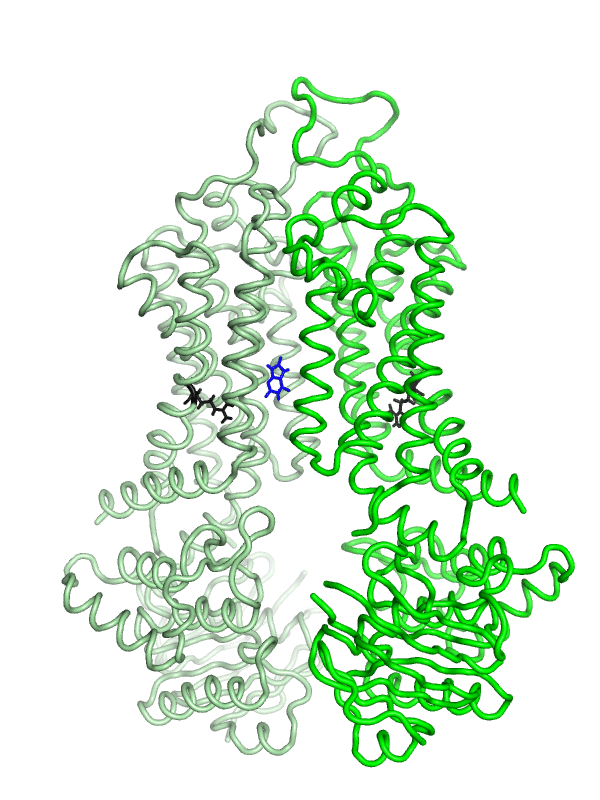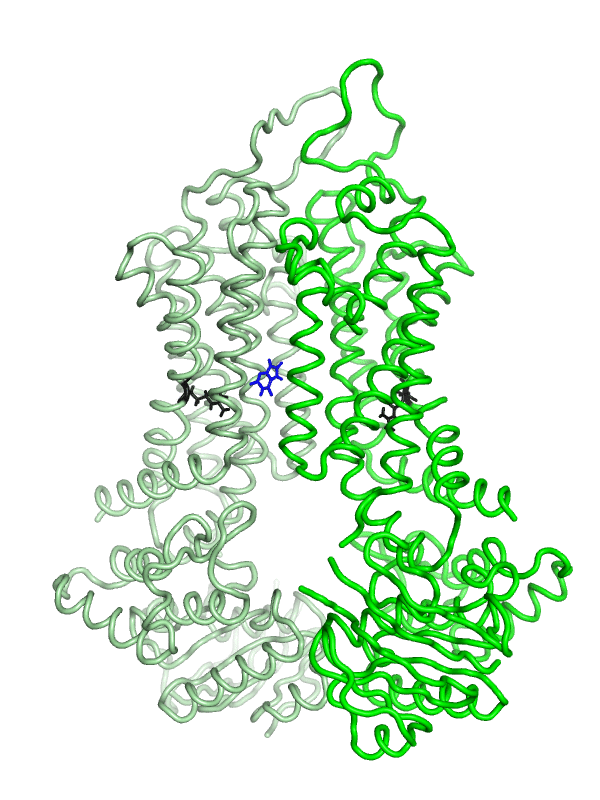
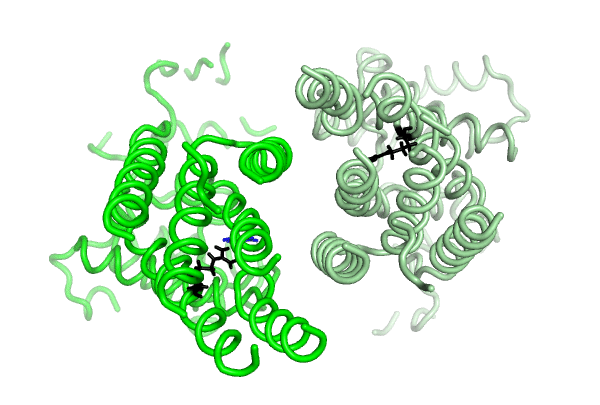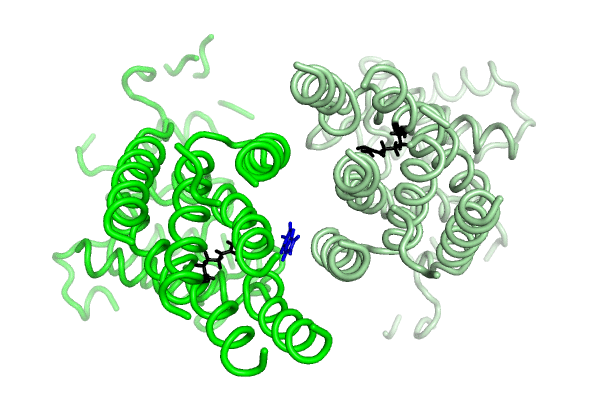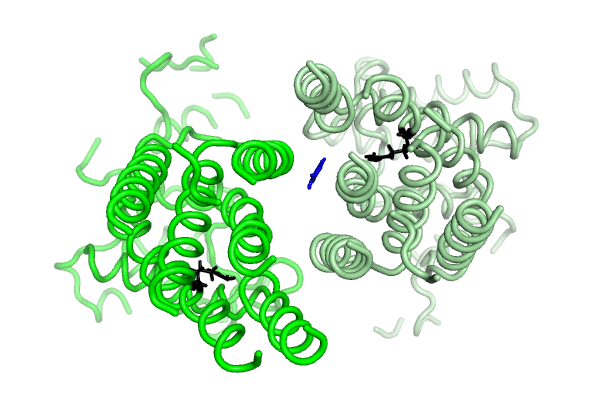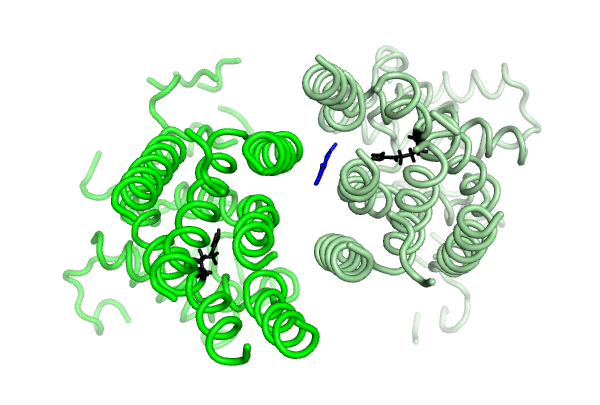
Equilibrium simulations show that uric acid, a substrate of ABCG2 visits Site 2, which was identified by Laszlo et al and involves R482:


| Homology model | This is the raw homology model (V1) used for input for CHARMM-GUI for the MD simulations described in our paper. It was generated using Modeller in two steps. In the second step the EL loops between between C592 and C608 were refined. In both steps one of the 100 models were selected based on DOPE score of Modeller. |
[protein PDB] [protein+POPC+water+ions PDB] [ABCG2/G5/G8 alignment FASTA] [All ABCG2 and CFTR NBD1 alignment FASTA] |
| This is the equilibrated homology model. The model V1 inserted in POPC bilayer with the same CHARMM-GUI setup as for MD, except that backbone atoms were restrained (4,000 kJ/mol/nm2) during the equilibration process. The last frame of a 10 ns production run (when the backbone atoms were also restrained) was selected. |
[protein PDB] [protein+POPC+water+ions PDB] |
|
 |
Reviewers make you think more and based on their notes we generated a second homology model (V2). We tried to model the linker region, but because of the longer sequence and low similarity we could insert only the first helix. We constrained these helices symmetrical in both ABCG2 halves, that corrected the distortion of this region in the ABCG8. Although we also modeled the "regulatory insertion" using Modeller, this structural segment should be also considered as a low quality region. We have information neither on secondary structural elements in this region nor on its spatial localization (e.g. if intercalated between the NBDs or not). We also built the extracellular loops differently compared to model V1, forcing their symmetry. The extracellular loop exhibits lower sequence similarity and their specific conformation in the model should be considered only one possible conformation because of its flexibility. |
[protein PDB] [protein+POPC+water+ions PDB] [visual PSE] |
| This is the equilibrated model V2. |
[protein PDB] [protein+POPC+water+ions PDB] |
|
| Molecular dynamics | Example parameter files for GROMACS from CHARMM-GUI output. |
[energy minimization MDP] [equilibration step #1 MDP] [production run MDP] |
| In silico docking | Molecules docked using Autodock Vina. | [sdf and mol2 files ZIP] |
| Box and other parameters. |
[parameters TXT] [visual PSE] |